- Research
- Open access
- Published:
OsnR is an autoregulatory negative transcription factor controlling redox-dependent stress responses inCorynebacterium glutamicum
188bet亚洲体育平台volume20, Article number:203(2021)
Abstract
Background
Corynebacterium glutamicumis used in the industrial production of amino acids and nucleotides. During the course of fermentation,C. glutamicumcells face various stresses and employ multiple regulatory genes to cope with the oxidative stress. TheosnRgene plays a negative regulatory role in redox-dependent oxidative-stress responses, but the underlying mechanism is not known yet.
Results
Overexpression of theosnRgene inC. glutamicumaffected the expression of genes involved in the mycothiol metabolism. ChIP-seq analysis revealed that OsnR binds to the promoter region of multiple genes, includingosnRand cg0026, which seems to function in the membrane-associated redox metabolism. Studies on the role of theosnRgene involving in vitro assays employing purified OsnR proteins and in vivo physiological analyses have identified that OsnR inhibits the transcription of its own gene. Further, oxidant diamide stimulates OsnR-binding to the promoter region of theosnRgene. The genes affected by the overexpression ofosnRhave been found to be under the control of σH.In theosnR-overexpressing strain, the transcription ofsigHis significantly decreased and the stimulation ofsigHtranscription by external stress is lost, suggesting thatosnRandsigHform an intimate regulatory network.
Conclusions
Our study suggests that OsnR not only functions as a transcriptional repressor of its own gene and of those involved in redox-dependent stress responses but also participates in the global transcriptional regulation by controlling the transcription of other master regulators, such assigH.
Background
Corynebacterium glutamicumis a Gram-positive microorganism and classified into the orderActinomycetales, which also comprises species such asMycobacteriumandStreptomyces[1].C. glutamicumis predominantly aerobic and commonly used for the industrial production of amino acids and nucleotides [2].During the course of fermentation, microorganisms encounter various intracellular and extracellular stresses, among which oxidative stress imposes a major challenge to cells [3,4].The responses ofC. glutamicumcells to stress-causing factors have been studied in some detail, and their molecular regulatory mechanisms are now being unveiled [5].
Reactive oxygen species (ROS), such as hydrogen peroxide (H2O2), are formed during aerobic respiration and can react with major cellular constituents, including DNA, lipids, proteins, iron-sulfur clusters, and the amino acids cysteine and methionine, in various ways, leading to cell damage [6,7].C. glutamicumcells are equipped with various enzymatic and non-enzymatic measures, such as catalase and mycothiol (MSH), respectively, that can cope with ROS and stress-caused impairments. InC. glutamicum, thekatAgene encodes the H2O2-detoxifying catalase, and OxyR acts as the main transcriptional repressor of thekatAgene [8,9].Peroxidases, which have higher affinities for H2O2than catalase, have also been detected inC. glutamicum[10].Low-molecular-weight thiols such as mycothiol (1-d-myo-inosityl-2-[N-acetyl-l-cysteinyl]amido-2-deoxy-α-d-glucopyranoside) function in the maintenance of the cellular redox homeostasis [11,12,13] by cycling between oxidized and reduced forms [14].Mycothiol disulfide reductase, encoded by themtrgene, catalyzes production of the reduced form of mycothiol at the expense of the reductant NADPH [15].Mycothiol has also been implicated in the detoxification of toxins and antibiotics [13,16,17].In addition, thioredoxin, a small protein, represents another prevalent thiol-based redox enzyme system and plays important roles in protecting proteins from oxidative damage [18,19].For example, reduced thioredoxin, which is produced by thioredoxin reductase (encoded bytrx), is involved in the repair of oxidized proteins through the cysteine thiol-disulfide exchange mechanism [20].Consequently, the cellular concentration of NAD(P)H is critical, because this biomolecule serves as the main source of reducing power for cellular factors, such as thioredoxin, mycothiol, and peroxidases [10,21].
Significant progress has been made in recent years on identifying the proteins that play regulatory roles in the oxidative-stress responses inC. glutamicum.Remarkably, numerous regulatory proteins participate in these responses. Moreover, many of these proteins contain cysteine residues in a configuration that may respond to cellular redox signals, thereby regulating cognate stress-responsive genes. These cysteine-containing regulators inC. glutamicuminclude WhcE and WhcA [22,23], OxyR [8,24], OhsR [25], RosR [26], MsrR [27], CosR [28], QorR [29], OasR [30], and OsrR [31].In addition, along with the master regulator SigH [32,33], multiple regulatory proteins also participate directly or indirectly in the regulation of oxidative-stress responses [34,35].
Recently, Jeong et al. [36] have found that theosnRgene plays a negative role in the oxidative-stress responses inC. glutamicumand suggested a role for this gene in redox-mediated stress-response systems. Additionally, theosnR-overexpressing strain shows retarded growth, decreased transcription of thetrxandmtrgenes, and sensitivity to oxidants, such as H2O2and diamide. However, the precise molecular function ofosnRis still unclear. Accordingly, this study aimed to unveil the role of theosnRgene at the molecular level.
Results
Overexpression of theosnRgene affects the mycothiol metabolism
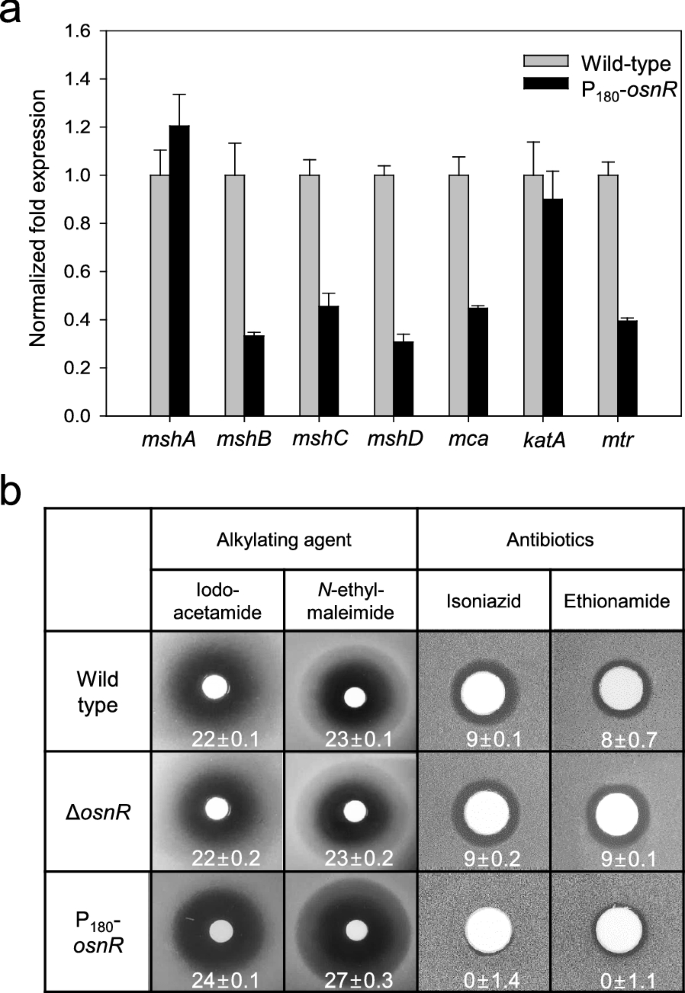
Previously, we have reported that theosnRgene plays a negative regulatory role for the genes involved in reductant-dependent ROS detoxification [36].Further, Jeong et al. [36] have found that theosnR-overexpressing strain (P180-osnR) shows an imbalanced NADPH/NADP+ratio and downregulated transcription of genes encoding thioredoxin reductase (trx) and mycothiol disulfide reductase (mtr), and thus postulated that the strain is deficient in the redox metabolism. Since MSH is known as the major low-molecular-weight thiol, playing important roles in protectingC. glutamicumcells from oxidative stress, we analyzed the transcription levels of genes involved in mycothiol biosynthesis in P180-osnRstrain. Genes, such asmshB(MSH deacetylase, cg1250),mshC(MSH ATP-dependent ligase, cg1709), andmshD(MSH acetyltransferase, cg2847) showed 55–70% decreased transcription compared with that of the wild type strain (Fig.1a), suggesting a deficient mycothiol metabolism in P180-osnRcells. Themcagene (MSHS-conjugated amidase, cg1127), which is involved in the regeneration of mycothiol from the mycothiol-mediated detoxification product [17], also showed decreased transcription. Unlike these genes,mshA(MSH glycosyltransferase, cg0481), which encodes the first enzyme of the five-step biosynthesis process, showed marginal transcriptional upregulation in P180-osnRstrain. Meanwhile, the transcription of the catalase-encodingkatA基因,作为阳性对照(36], was only marginally affected (Fig.1a).
The mRNA levels of mycothiol biosynthetic genes inC. glutamicumcells and the sensitivities of theC. glutamicummutants to alkylating agents and antibiotics.aC. glutamicumwild-type andosnR-overexpressing (P180-osnR) cells were grown in the minimal medium, and the mRNA levels were measured using qRT-PCR as described in “Materials and methods” section. Error bars indicate the standard deviation of three replicates from a representative experiment.bPaper discs placed on MB plates containing lawns ofC. glutamicumwild-type,osnR-deleted (ΔosnR), or P180-osnRcells were spotted with iodoacetamide,N-ethylmaleimide, isoniazid, or ethionamide. Diameters of the zone of inhibition are shown in millimetres
To test whether the altered transcription levels of mycothiol biosynthetic genes in P180-osnRstrain decreased the cellular concentration of mycothiol, we compared the sensitivity of P180-osnRcells to alkylating agents with that of the wild-type cells because intracellular mycothiol is known to be involved in the detoxification of these compounds [13,16].As shown in Fig.1b, P180-osnRcells showed an increased sensitivity to the thiol-attacking alkylating agents, such as iodoacetamide andN-ethylmaleimide, suggesting a diminished level of mycothiol in this strain. No significant difference in sensitivity was observed between the wild type and ΔosnRstrains. Further, we tested the response of P180-osnRstrain to antibiotics, such as isoniazid and ethionamide, which are pro-drugs that need to be activated by mycothiol [37,38].As expected, P180-osnRstrain showed significant resistance to the antibiotics (Fig.1b), consistent with a diminished level of intracellular mycothiol [12].Collectively, these data show that the expression of the genes involved in the mycothiol metabolism might have been affected by the overexpression ofosnR, whereby the cytosolic redox homeostasis involving mycothiol is disturbed.
Identification of potential targets of OsnR via ChIP-seq
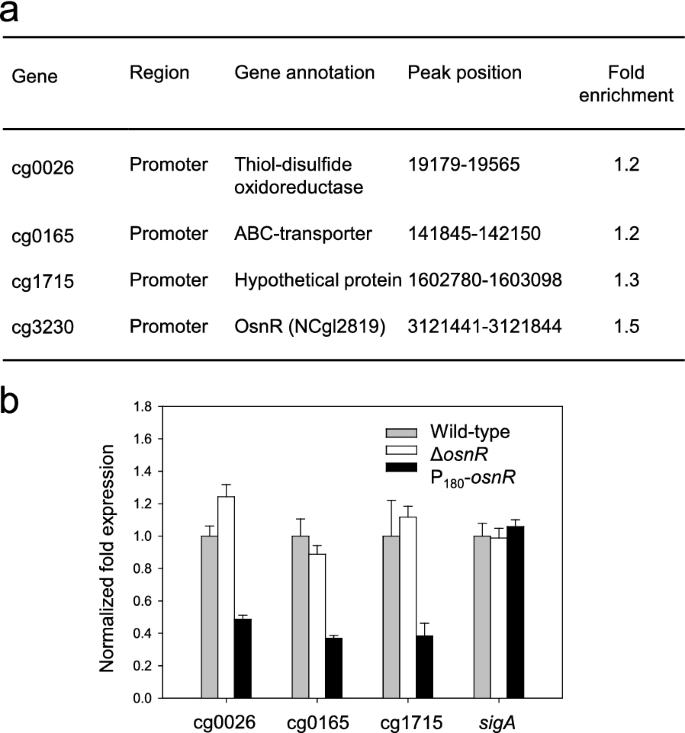
Because OsnR affected the transcription of various genes involved in redox reactions [36一个nd the mycothiol metabolism (Fig.1a), we postulated thatosnRhas a general regulatory role in the redox metabolism. Therefore, we performed a ChIP-seq analysis using HL1653 strain, which over-expresses the Myc-tagged OsnR protein (Myc-OsnR), to identify direct chromosomal targets of the OsnR protein. Because specific antibodies against the OsnR were not commercially available, we used a Myc-tagged OsnR protein for the analysis. As shown in Fig.2a, although only a few target sites were identified, all of them were located in the promoter or regulatory regions of genes, including cg0026, cg0165, cg0175, and cg3230 (osnR), suggesting a transcriptional regulatory function of the OsnR protein. Although the values of fold enrichment were rather low, the identified targets passed the threshold point, indicating that the values are statistically meaningful. Interestingly, the promoter region ofosnRwas identified as the most prominent target of the OsnR protein. The other targets included the promoter regions of cg0026 (thioredoxin domain-containing protein) and cg0165 (ABC-type transporter) genes. In particular, cg0026 encoded a protein that showed 42.8% identity to the DsbA-like thiol-disulfide oxidoreductase ofStreptomyces coelicolor, which seems to function in the membrane-associated redox metabolism [39].In accordance with the ChIP-seq data, the transcription of the identified genes cg0026, cg0165, and cg1715 was notably decreased in P180-osnRstrain (Fig.2b).
Target sites of the OsnR protein, as identified through the ChIP-seq analysis, and transcription levels of the identified genes inC. glutamicumcells.aThe table shows the binding region of the Myc-tagged OsnR protein on the chromosomal DNA, as identified through the ChIP-seq analysis (see the “Materials and methods” section for the experimental details).bqRT-PCR analysis of the identified genes inC. glutamicumcells. The mRNA levels in cells grown in the minimal medium were measured. Three independent experiments were performed, and the data represent three technical replicates from a representative experiment
OsnR directly binds to the promoter ofosnR
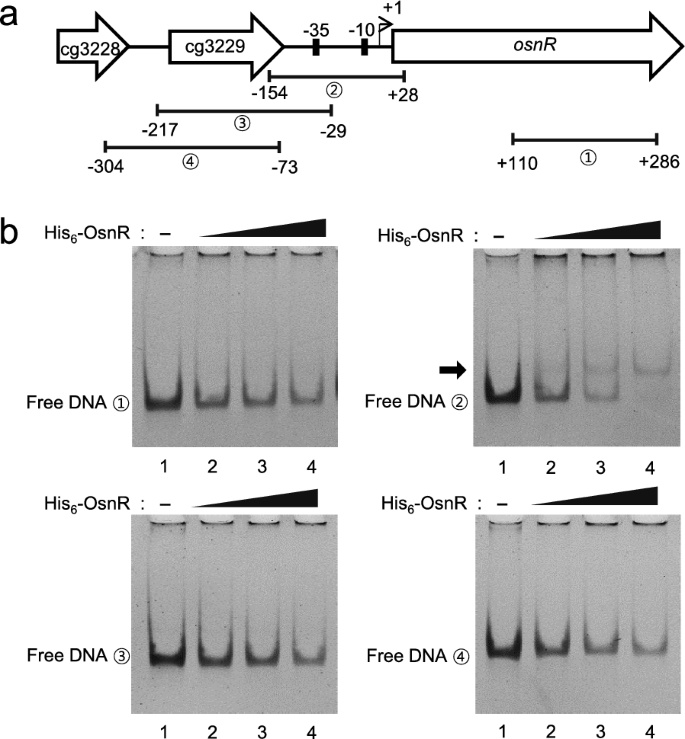
Based on the ChIP-seq data, we assessed for DNA–protein interaction in a purified system. The overexpressed OsnR protein was purified in the form of histidine-tagged protein (His6-OsnR), which was obtained from inclusion bodies after the on-column refolding process. As shown in Fig.3b, electrophoretic mobility shift assays (EMSA) showed binding of the His6-OsnR启动子区域(从154−+ 28)of its own gene, as evidenced by the shifted bands, whereas DNA involving the upstream region of the promoter (from − 217 to − 29) did not, suggesting that the binding sites are located in the DNA region spanning from − 29 to + 28 (the transcriptional start site (+ 1) was based on published data [40]). The location of the presumed binding site is in accordance with the findings of Jeong et al. [36], who reported the negative regulatory role of the OsnR protein. As expected, DNA fragments containing either the promoter region of the upstream gene cg3229 or the coding region of theosnRgene (from + 110 to + 286) did not show any shifted band (Fig.3b).
Binding of the purified OsnR protein to the promoter region of theosnRgene.aSchematic diagram of the chromosomal region comprising theosnRand adjacent genes. Numbers in circles indicate the DNA fragments used in the EMSA.bEMSA was performed using the purified His6-OsnR protein and the DNA fragments shown ina.The reactions in lanes 1, 2, 3, and 4 contained 0, 0.8, 1.0, and 1.2 μg of the OsnR protein, respectively. The arrow indicates the shifted band
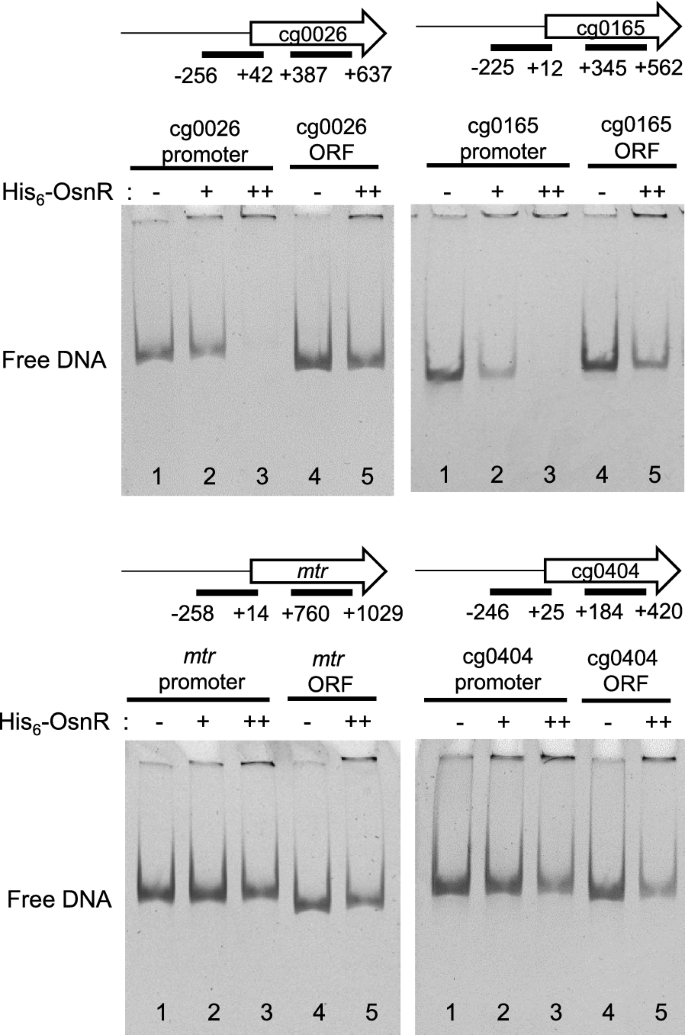
We further analyzed the binding of the OsnR protein on additional promoters identified in the ChIP-seq analysis. As shown in Fig.4, although band shifts were not evident with the promoter regions of the cg0026 (from − 256 to + 42) and cg0165 (from − 225 to + 12) genes, free DNA clearly disappeared in the presence of His6-OsnR, suggesting a DNA–protein interaction. The disappearance of free DNA was not observed with DNA fragments containing the internal coding region of the genes. We also tested the binding of OsnR on the promoter region of additional genes, whose expression was affected by the overexpression ofosnR[36].No DNA-OsnR interaction was observed with the promoter region of themtr(from − 258 to + 14) and cg0404 (a nitroreductase-family protein, from − 246 to + 25) genes [36].In addition, OsnR was not observed to bind to the promoters oftrxBandsodA, either (Additional file1: Fig. S1). Overall, these data show that OsnR functions as a DNA-binding transcriptional factor, which may also repress its own transcription.
The DNA-binding activity of OsnR is redox-dependent
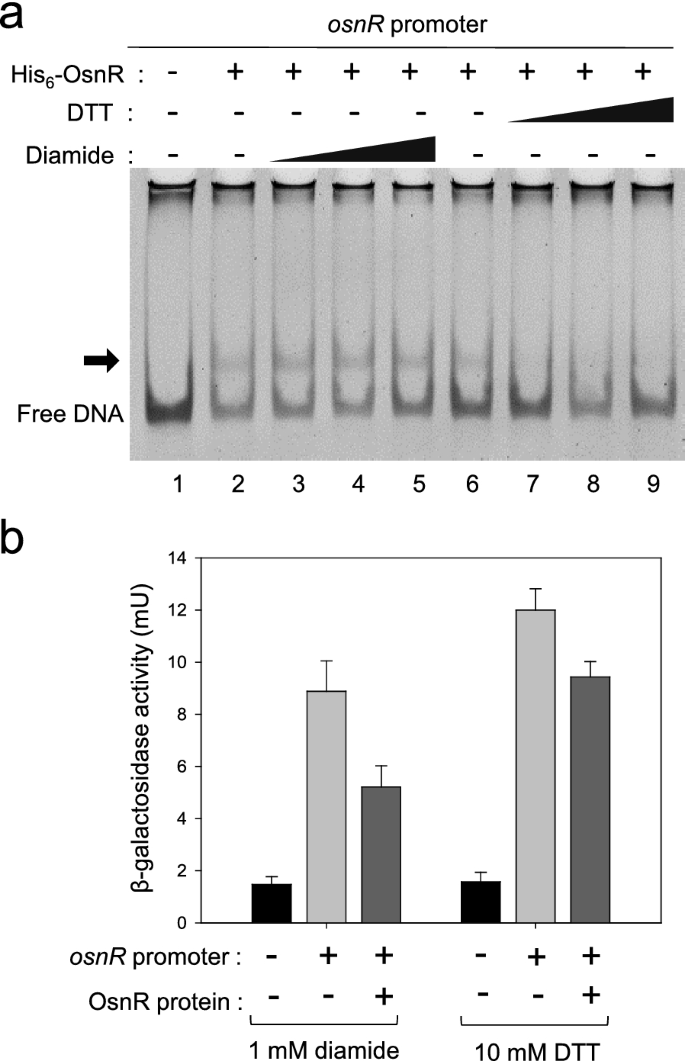
Based on the data that OsnR may play an autoregulatory role for the transcription of its own gene, we tested the effect of the redox status of the reaction mixture on the DNA-binding activity of the protein. As shown in Fig.5a, the addition of the oxidizing agent diamide stimulated OsnR binding to the promoter region spanning from − 154 to + 28. However, when diamide was replaced with DTT, not only the stimulatory effect but also the shifted bands disappeared. This observation suggests that OsnR binds to the regulatory region of its own gene in cells exposed to oxidative stress, thereby inhibiting the transcription of the gene. To test whether the binding of OsnR represses its own transcription, we designed an in vivo experiment as described below. First, we constructed a reporter plasmid carrying theosnRpromoter fused to the upstream region of thelacZgene. Next, the constructed plasmid pSL553-PosnR::lacZwas introduced intoE. colicells along with plasmid pKK223-3-Ptac::osnR.In the resulting strain, the OsnR protein could be induced with IPTG, and the binding of the expressed OsnR protein to theosnRregulatory region could be monitored by measuring the β-galactosidase activity. When cells carrying the reporter plasmid pSL553-PosnR::lacZalong with the empty vector pkk223-3 were grown with DTT, approximately 12 mU of β-galactosidase activity was observed, indicating that theosnRpromoter is recognized by theE. colitranscription apparatus. The introduction of plasmid pKK223-3-Ptac::osnRinto theE. colistrain carrying the reporter plasmid and subsequent expression of the OsnR protein resulted in approximately 20% reduction in the β-galactosidase activity (Fig.5b). Conversely, cells grown in the presence of diamide showed 40% reduction in the β-galactosidase activity. These data suggest that the binding of the OsnR protein to the regulatory region of theosnRgene is stimulated by diamide, whereby the expression of thelacZgene is decreased. Collectively, these data suggest that OsnR functions as a transcriptional repressor and the DNA-binding activity of the OsnR protein is modulated by cellular redox status.
Effects of diamide and DTT on the DNA-binding activity of OsnR.aEMSA assay showing the effect of diamide (lanes 3–5) or DTT (lanes 7–9) on the DNA-binding activity of the OsnR protein. The binding of the purified His6-OsnR on the promoter region ofosnRwas assayed using EMSA. The experimental details are shown in the Material and Methods section.bAn in vivo β-galactosidase assay showing the binding activity of OsnR on the promoter region of theosnRgene. The binding activity was assessed by measuring the β-galactosidase activity expressed from the PosnR::lacZreporter plasmid. TheE. colihost contained a plasmid that expresses the OsnR protein and the reporter. Cells were grown in the presence of DTT or diamide.The shown β-galactosidase activity is the mean value from at least three independently performed experiments
Global transcriptional regulatory role of OsnR
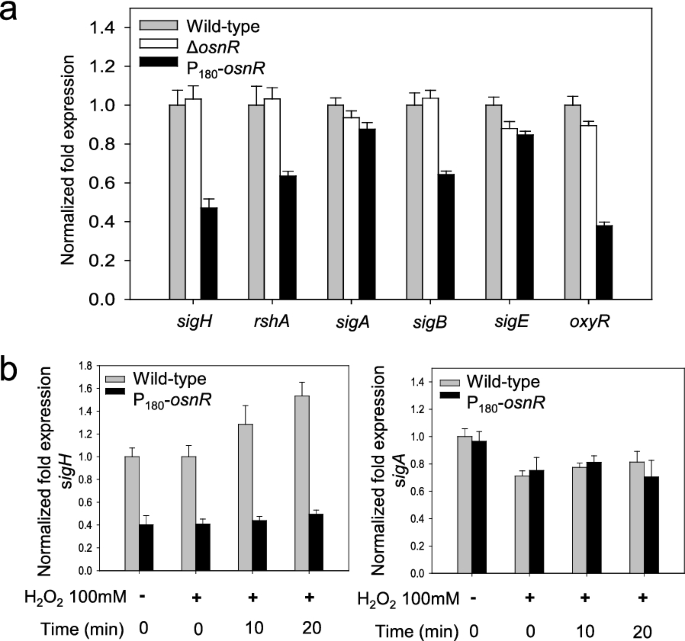
Because OsnR did not show direct binding to the promoter regions of the oxidative-stress-responsive genesmtrandtrx, which showed decreased transcription in P180-osnRstrain, we postulated thatosnRmay play an indirect regulatory role, conveying its activity through other regulatory genes. Moreover, the severe growth impairment of P180-osnRstrain and the down-regulation of redox-responsive genes in the strain [36一个lso suggested a role of theosnRgene in a global regulatory cascade. Among the master regulatory genes, which might be involved in the regulation of stress-responsive genes, we chose thesigHgene (cg0876), which not only plays a major regulatory role in stress responses but is also involved in the transcription oftrx,mtr, and mycothiol metabolic genes [33,41,42].We also observed that thesigH-regulatedftnanddpsgenes, which are implicated in the iron homeostasis [43,44], were down-regulated in P180-osnRstrain (Additional file1: Fig. S2). Based on this knowledge, we studied the transcription ofsigHin the ΔosnRandosnR-overexpressing P180-osnRstrains. As shown in Fig.6a, the transcription level of thesigHgene in P180-osnRstrain was at 50% of that observed in the wild type and ΔosnRstrains. The genershA, which constitutes an operon withsigH[33,41一个nd encodes an anti-sigma-factor, showed decreased transcription in P180-osnRstrain. ThershAgene is also known to have its own promoter, which is σH-dependent. Conversely, the expression level ofsigA, encoding a housekeeping sigma factor, was only marginally affected in P180-osnRstrain, whereas that ofsigB, which encodes a primary-like sigma factor σBand plays roles in the transition phase between the exponential and stationary growth phases, was decreased by approximately 30% of that of the wild type strain (see below). These results suggest that transcriptional changes observed in many oxidative-stress response genes in P180-osnRstrain might be in part due to decreased expression ofsigH,which constitutes the large group of the σHregulon, of which many genes, includingsigB, are members [42,45].Next, we measured the transcription level of the regulatory geneoxyR(cg2109), which acts as a transcriptional repressor for thekatAgene. As shown in Fig.6a, theoxyRgene was transcribed only at a 40% level in P180-osnRcells, compared with the wild-type level. This observation agrees with the findings of Jeong et al. [36], who reported the de-repression ofkatAin P180-osnRstrains.
The transcription levels of σ-factor–encoding genes inC. glutamicumcells, and the effect of the hydrogen peroxide on the transcription ofsigH.aC. glutamicumwild-type,osnR-deleted (ΔosnR), andosnR-overexpressing (P180-osnR) cells were grown in the minimal media, and the mRNA levels were measured using qRT-PCR.bThe transcription levels ofsigHandsigAinC. glutamicumwild-type and P180-osnRcells after challenging the cells with hydrogen peroxide. Error bars indicate the standard deviation of three replicates from a representative experiment
To elucidate the regulatory interaction between theosnRandsigHgenes, we performed a physiological analysis as described below. First,C. glutamicumcells were grown to the mid-exponential growth phase in the minimal medium and were challenged with H2O2, which can oxidize protein thiols [46].Next, the cells were harvested at appropriate time points, and the cellularsigHmRNA levels were quantitated via qRT-PCR. As shown in Fig.6b, the H2O2challenge caused increased transcription ofsigHin wild-type cells. Stimulation ofsigHwas not observed in OsnR-overexpressing P180-osnRcells, and the level of transcription was at 40% of that of the wild-type cells. However, the transcription ofsigAwas almost identical in the wild-type and P180-osnRstrains regardless of the H2O2challenge. Collectively, these data suggest that theosnRgene may regulate the transcription ofsigH.
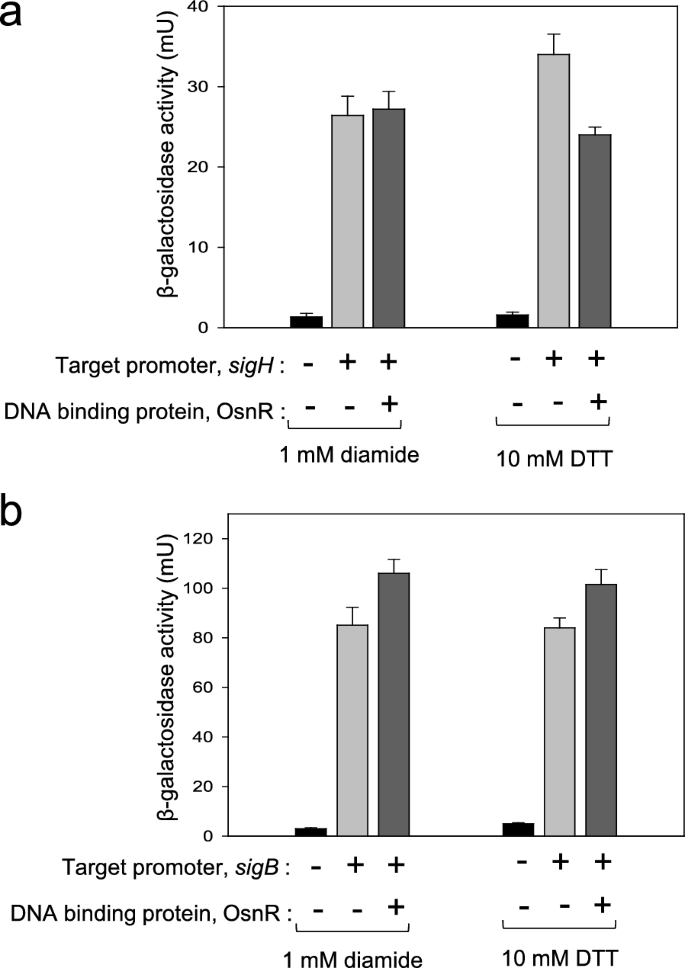
As the next step, we performed EMSA with the purified OsnR protein. As mentioned in the earlier section, the protein was purified in the form of His6-OsnR through the on-column refolding process. Although there were signs of protein binding on the regulatory region ofsigH, it was not evident (see “Discussion”). Suspecting a weak interaction between the protein and target DNA, we switched to in vivo assays to quantitatively monitor the interaction. First, we constructed a reporter plasmid carrying thesigHpromoter fused to thelacZgene to monitor thesigHpromoter activity through β-galactosidase activity. Next, the constructed plasmid pSL553-PsigH::lacZwas introduced intoE. colicells along with plasmid pKK223-3-Ptac::osnR.Subsequently, the binding activity of the expressed OsnR protein to thesigHregulatory region was quantitated in cells grown with DTT or diamide. As shown in Fig.7a, the presence of OsnR in cells grown in the presence of the reductant DTT repressed the β-galactosidase activity by 30%, suggesting that the OsnR protein bound to the regulatory region of thesigHgene. However, when cells were grown in the presence of diamide, the repression of thesigHgene was not observed. Next, the binding of the OsnR protein to the promoter region of thesigBgene, which also showed decreased transcription in P180-osnRcells, was tested through the constructed plasmid pSL553-PsigB::lacZ.In contrast to the findings regarding thesigHgene, no significant reduction of β-galactosidase activity was observed in cells grown with DTT or diamide (Fig.7b), indicating that the interaction between the regulatory region ofsigHand OsnR is specific. Collectively, these data suggest that cells exposed to oxidative stress initiate the transcription of thesigHgene by eliminating the repression exerted by the OsnR protein on the regulatory region ofsigH.
In vivo assays showing the binding of the OsnR protein to the promoter region of thesigHgene. The binding activity was analyzed by measuring the β-galactosidase activity expressed from the PsigH::lacZreporter plasmid (a) and the PsigB::lacZreporter plasmid (b). TheE. colihost contained a plasmid that expresses the OsnR protein and the reporter. A plasmid carrying only the reporter was used as a negative control (b). The cells were grown with DTT or diamide. Three independent experiments were performed, and the data represent three technical replicates from a representative experiment
Discussion
TheosnRgene has been suggested to play a global regulatory role as well as a negative role in redox-associated stress responses [36].In this study, we demonstrated its DNA-binding activity as a transcriptional repressor for its own gene and cg0026, which is presumably involved in the redox-metabolism. The binding of the OsnR protein to the regulatory region of its own gene was evident, although weak interactions with other promoters were observed as well. Overexpression of the protein inE. coliand refolding process, which were applied to obtain pure OsnR protein, might be attributed to the low activity of the protein. Further, considering the small size of the protein (127 amino acids), histidine-tagging (His6-OsnR) may have hindered the interaction between OsnR and target DNA fragments. Nevertheless, weak binding of OsnR to the targets, such assigHregulatory region, may be an intrinsic property of the interaction because such a property can be beneficial to cells, when one critically considers the inhibitory role of theosnRgene in cell physiology [36].For instance, since thesigHgene is involved in the regulation of a broad spectrum of cellular activities, active regulation ofsigHby a strong interaction with OsnR, whose role is confined to responses involving the redox homeostasis, may be physiologically inadequate to cells.
The binding of the purified OsnR on the regulatory region of its own gene shows that theosnRgene is autoregulated. Autoregulation of theosnRgene may suggest its critical role in cell physiology, because autorepression, known as one of the most efficient regulatory mechanisms [47),可能拯救细胞变化的响应时间environmental conditions. Similarly, thesigHgene is also known to be autoregulated by its own gene product [33,48].The OsnR binding site, located between − 29 and + 28 of theosnRgene, indicates that the OsnR protein functions as a repressor. Stimulation of the binding of OsnR to its own promoter in the presence of the oxidant diamide agrees well with previous reports, which showed a negative regulatory role of the gene [36].Logically, cells exposed to diamide will downregulate OsnR, due to transcriptional repression of theosnRgene by its own protein, thereby de-repressing the stress-responsive genes. In addition, changes in cellular redox status can cause structural modification of the OsnR protein through cysteine residues, resulting in changes in the activity and functional properties of the protein, as such mechanism was proposed for other regulators, such as CosR (C49 and C62), MsrR (C62), OasR (C95), OhsR (C125), OxyR (C199 and C208), RosR (C92), and QorR (C17) [25,26,27,28,29,30,49,50,51].The OsnR protein has two cysteine residues (C2 and C10) at the N-terminus, and these residues may function as a thiol-based redox switch to respond to cellular redox status. Further, since the OsnR protein is predicted to form a homo-dimeric protein through the N-terminus, its dimerization capability could be modulated by cellular redox status.
Identification of only a handful of genes via the ChIP-seq analysis may suggest a global role of theosnRgene, which may exert its regulatory effect through other master regulators, such assigH.However, as mentioned above, the intrinsically weak DNA–protein interaction might have resulted in unveiling only a few target genes. The growth condition employed for the assay may not be ideal for the OsnR protein to bind to target DNAs. In addition, the Myc-tagging of the OsnR protein may have lowered the binding activity of the protein, thus diminishing the number of identified target genes. The OsnR protein contains a helix-turn-helix DNA-binding motif at the C-terminus. Although the Myc-tag at the N-terminus worked well as an epitope for the binding of the 9E10 antibody, the tagging might have altered the protein conformation, thereby altering the dimerization capacity and, subsequently, the DNA-binding ability of the protein. Indeed, such negative effects have been reported in studies involving ChIP-seq [52,53,54,55].
Although we do not have a clear picture of how theosnRgene exerts its regulatory role inC. glutamicum, it is evident that it functions in the gene network involving the ECF-type sigma factor σH, which primarily functions in heat- and oxidative-stress responses [22,41].The in vivo assays (Fig.7) showed that the repression ofsigHby OsnR is relieved in cells challenged with diamide, indicating a close regulatory interaction betweenosnRandsigH.The findings by Kim et al. [32], which showed regulated expression of thesigHgene, suggest the involvement of transcriptional factors, among which OsnR may be one of them. Likewise, the LexA repressor, which is involved in the regulation of the SOS response inC. glutamicum, also seems to participate in the regulation of thesigHgene, as suggested by the presence of a SOS box in the regulatory region of thesigHgene [33,56].
The transcriptional downregulation of the mycothiol metabolic genesmshB,mshC,mshD, andmcain theosnR-overexpressing cells seems to be caused by the low intracellular level of σH, because these genes have been reported to be part of the σHregulon [17,33,57,58].This conclusion is further supported by the fact that themshAgene, which did not show notable transcriptional changes in P180-osnRcells, is not a member of thesigHregulon inC. glutamicum[59,60].ThesigHgene is known to occupy the central position in the cross-regulated network of sigma factors in controlling genes involved in various stress responses, even the SOS response [29,32,33,41,57].The general down-regulation of redox-responsive genes in P180-osnRstrain could be due to a subordinate regulatory effect, as cross-regulation among sigma factors are frequent, and multiple sigma factors are often involved in the regulation of some genes [42,61].The general growth defect of P180-osnRstrain [36] may support this idea. Demonstration of the OsnR binding to the regulatory region ofsigHis necessary to demonstrate the role of OsnR in the transcription of thesigHgene.
The identification of cg0026 as one of the targets of OsnR in the ChIP-seq, and the binding of OsnR to the regulatory region of cg0026 (Fig.4) is noteworthy. These observations indicate that theosnRgene specifically regulates genes involved in cellular redox reactions and globally participates in stress responses throughsigH.The cg0026 gene has been annotated to encode a secreted protein containing a thioredoxin domain (i.e. thiol-disulfide oxidoreductase) and forms an operon with cg0025. The cg0025 gene encodes an integral membrane protein, which shows 27–33% identity with the cytochrome-biogenesis protein CcdA. Protein disulfide-isomerases, such as thiol-disulfide oxidoreductase, play important roles not only in the formation of disulfide bonds but also in reducing incorrect disulfide linkages [39].Protein folding in the outer vicinity of the cell membrane is challenging due to the oxidative nature of the environment. InEscherichia coli, DsbA, which harbors a thioredoxin-like fold [62], catalyzes the formation of disulfide bonds in unfolded proteins as they are secreted into the periplasm. The DsbA active site involves a reactive disulfide bond found in a CXXC consensus sequence [62].DsbA is regenerated by the membrane protein DsbB, which exchanges the disulfide bonds [63,64].Additionally, damaged periplasmic proteins by oxidative stress can be repaired by the periplasmic DsbC and membrane-bound DsbD protein pair [39].Furthermore, CcdA, which is a functional homolog of DsbD, provides reducing equivalents for the reduction of cytochrome c.Corynebacteriumspecies secrete cysteine-containing proteins into the exoplasm [65,66].InC. diphtheria, the membrane-localized disulfide-bond-forming reactions are catalyzed by MdbA and VKOR-like proteins, which are DsbAB-like proteins [39].However, the proteins encoded by cg0026 and cg0025 do not show significant homology to the proteins, suggesting a distinctive role of the cg0026-encoded protein inC. glutamicum.Considering the role ofosnRin responses involving oxidative stress, and the formation of a single transcriptional unit with cg0025, it is logical to speculate that the cg0026-encoded protein may play a role analogous to that of the DsbC ofE. coli.Further studies are necessary to elucidate whether the cg0026 protein is involved in repairing non-native disulfide bonds in the exoplasm.
Conclusions
We found that theosnRgene specifically regulates genes involved in redox-dependent stress responses. OsnR functions as a transcriptional repressor and responds to cellular redox status. TheosnRgene may also participate in global transcriptional regulation through other regulators, such assigH,这是主人的监管作用in cells exposed to heat or oxidative stress. This work provides a deeper understanding of the redox-dependent stress-responsive regulatory networks ofC. glutamicum.
Materials and methods
Bacterial strains, plasmids, and culture conditions
All the strains and plasmids used in this study are listed in Additional file1: Table S1.C. glutamicumAS019E12 was used as the wild-type strain.C. glutamicumHL1638 and HL1643 were used as the ΔosnRmutant andosnR-overexpressing strains, respectively.C. glutamicumHL1653 harbours plasmid pSL580, which expresses the Myc-tagged OsnR protein (Myc-OsnR).E. coliDH10B (Invitrogen) was used to construct and propagate the plasmids.E. coliBL21 DE3 (Merck) was used for the expression of the His6-tagged OsnR protein (His6-OsnR).E. coliandC. glutamicumstrains were cultured in Luria–Bertani (LB) broth at 37 °C and MB medium at 30 °C, respectively [67,68].MCGC minimal medium with 1% (wt/vol) glucose was prepared as described previously [36].Antibiotics were added at the following concentrations: 50 μg/ml ampicillin, 25 μg/ml kanamycin, and 10 μg/ml tetracycline.
Construction of plasmids
Standard molecular cloning and transformation methods were employed [68].质ids were introduced intoC. glutamicumcells via electroporation [67].Restriction enzymes and DNA-modifying enzymes were used according to the manufacturer’s instructions (Takara Bio). PCR amplification of DNA fromC. glutamicumAS019E12 chromosome was performed using the primers listed in Additional file1: Table S2.
The plasmid expressing Myc-OsnR was constructed as follows: first, primers carrying the sequences needed for the amplification of theosnRgene were designed and additional sequences, which can optimally express the Myc epitope (5′-EQKLISEEDL-3′) inC. glutamicum.The resulting primers myc_osnRF and myc_osnRR (Additional file1: Table S1) were used to amplify the chromosomalosnRgene. Following the amplification, the PCR product was digested with PstI and inserted into the PstI site of pSL360 [69].The resulting plasmid pSL580, which expresses Myc-OsnR, was then introduced intoC. glutamicumto generate strain HL1653. Over-transcription of the DNA encoding OsnR-Myc was verified by the RT-qPCR analysis (Additional file1: Fig. S3). The pSL581plasmid expressing His6-OsnR was constructed by amplifying theosnRgene by using primers pET28a_osnRF and pET28a_osnRR, digesting the PCR product with NdeI and EcoRI, and subsequently inserting the resulting fragment in between the NdeI and EcoRI sites of pET28a vector (Novagen).
质id pSL592, which can express theosnRgene inE. coli, was constructed by amplifying theosnRgene by using primers pKK223-3_osnRF and pKK223-3_osnRR, digesting the PCR product with EcoRI and PstI, and inserting the resulting fragment into pKK223-3 vector (Amersham Pharmacia). The reporter plasmid carryinglacZwas constructed as follows: first, the region of pRS415 [70] that contains the T14, multiple cloning site, andlacoperon was amplified using primers pRS415_F and pRS415_R. Subsequently, the PCR product was digested with ScaI, and the resulting fragment was inserted into pACYC184 vector (New England BioLabs), generating plasmid pSL553. Next, appropriate promoters were introduced into pSL553 as follows: the promoter and regulatory regions were amplified using primers pSL553_osnRF/pSL553_osnRR, pSL553_sigHF/pSL553_sigHR, and pSL553_sigBF/pSL553_sigBF, digested with SmaI, and inserted into pSL553 to generate plasmids pSL594 (PosnR::lacZ), pSL595 (PsigH::lacZ), and pSL596 (PsigB::lacZ), respectively. The orientations and identities of the inserts were verified via DNA sequencing (Macrogen, South Korea).
RNA analysis
Corynebacterium glutamicumstrains were grown in MCGC minimal media and harvested at the early stationary phase. When necessary, H2O2was added to the cells during the mid-exponential growth phase to a final concentration of 100 mM, followed by 10–20 min incubation. After collecting the cells, their total RNA was extracted using the Nucleospin RNA II columns (Macherey–Nagel), and cDNA was synthesized using the ReverTra Ace qRT Kit (Toyobo). A CFX96 Real-Time PCR Detection System (Bio-Rad) was used as previously described [36,71].Reactions were performed in triplicate and relative ratios were normalized using the value for 16S rRNA. The primers used for qRT-PCR are listed in Additional file1: Table S2.
Physiological and biochemical analyses
Agar-diffusion assays were conducted as described previously [36,71].Lawn cells were mixed with 0.8% (v/v) top agar and poured onto MB plates. Paper disks (6.0 mm, Whatman), which were placed on the plates, were applied with the alkylating agent (10 μl of 100 mM iodoacetamide orN-ethlymaleimide) or antibiotic (200 mg isoniazid or ethionamide). The plates were photographed after 24 h of incubation at 30 °C.
Experiments involving measurement of β-galactosidase activity were performed as follows:E. colicells carrying appropriate plasmids were grown in LB medium at 37 °C and treated with 0.2 mM IPTG at the OD600of 0.5. The cells were then immediately treated with diamide or DTT and incubated at 30 °C for 3 h. Afterward, they were harvested, resuspended with the reaction buffer (5 mM Tris–HCl, 10 mM KCl, and 0.25% glycerol, pH7.5), and homogenized using the FastPrep-24 system (MP biomedicals). The supernatant obtained after centrifugation at 11,000×g5分钟是用作粗提取液。β的加actosidase assay was performed according to the report by Miller [72].One unit of activity was defined as the amount of enzyme that hydrolyzed 1 μmol of ONPG in 1 min at 30 °C. The protein concentrations were determined using the Bradford assay with bovine serum albumin solutions as the standard solutions [73].
ChIP-seq analysis
ChIP-seq analysis was performed by following the published methods [74] but with modifications. Myc-OsnR, constructed as described in the previous section, was used as bait to enrich the DNA segments with bound Myc-OsnR. Next, wild-typeC. glutamicumcells orC. glutamicumHL1653 cells expressing Myc-OsnR were cultured in 100 ml MCGC medium at 30 °C to the final OD600of ~ 2.0. Samples of 10 ml were collected and immediately mixed with formaldehyde (Extra Pure grade, Duksan) to give a final concentration of 1%, and incubated at 30 °C for 20 min with gentle agitation to induce DNA–protein cross-links. Subsequently, glycine was added to the final concentration of 125 mM, followed by incubation at room temperature for 5 min. The cells were harvested via centrifugation at 1600×gfor 10 min and washed twice with ice-cold phosphate-buffered saline (pH 7.4). They were then resuspended in 0.5 ml lysis buffer composed of 1% sodium dodecyl sulfate, 1% Triton X-100, 10 mM EDTA, 50 mM Tris–HCl (pH 8.1), 1 mM PMSF, and 5 μg/ml RNase A, incubated at 30 °C for 10 min, and then chilled on ice. The lysate was subjected to sonication (microprobe diameter of 3 mm; Sonics & Materials, Inc.) for 20 s at 30% amplitude on ice. The sonication process was repeated 10 times with 30 s interval to obtain chromosomal DNA fragments of 200–500 bp. Cell debris was removed via centrifugation at 11,000×gfor 10 min. The supernatant (5 μl) was collected and stored at − 80 °C for later use as the control input DNA. The rest of the cell extract was subjected to immunoprecipitation by using the Pierce Agarose ChIP kit (26156, Thermo Fisher Scientific). To immunoprecipitate the Myc-OsnR-bound DNA, 7 μg of c-Myc monoclonal antibody (9E10, Thermo Fisher Scientific) was added to the extract, and the mixture was incubated overnight at 4 °C. Then, 20 μl protein A/G and agarose beads (Thermo Fisher Scientific) were added to the mixture, followed by incubation at 4 °C for 2 h. The beads were washed twice with the wash buffer and resuspended with 150 μl elution buffer. The mixture was incubated at 65 °C for 30 min with shaking. The immunoprecipitated and control input DNA samples were diluted with the elution buffer and then treated with 20 μl proteinase K solution for 2 h at 65 °C. The samples were then purified using DNA clean-up columns (Thermo Fisher Scientific), precipitated, and resuspended in water. The resulting DNA samples were sequenced by Macrogen (South Korea) by using the HiSeq 4000 Sequencing System. All sequencing data have been deposited in ArrayExpress (accession number E-MTAB-11048).
Purification of His6-OsnR and EMSA
Escherichia coliBL21 (DE3) cells (Merck Bio-science) carrying pSL581, which overexpress His6-OsnR, were cultivated in LB medium. Recombinant-protein expression was induced by the addition of 0.2 mM IPTG at the OD600of 0.4. After cultivation for 4 h at 30 °C, the cells were harvested via centrifugation at 6000×gfor 10 min, resuspended in 10 ml buffer (20 mM HEPES, 500 mM NaCl, 40 mM imidazole, 8 M urea, 0.5% Tween 20, and 5% glycerol, pH 7.4), and lysed via sonication. The lysate was centrifuged at 11,000×gfor 1 h to remove cell debris. The resulting supernatant was filtered through a 0.2 μm syringe filter (Sartorius Stedim) before loading to a HisTrap FF column (GE Healthcare). Refolding of His6-OsnR was induced by applying 20 ml refolding buffer (20 mM HEPES, 500 mM NaCl, 40 mM imidazole, 0.5% Tween 20, 5% glycerol, and 5 mM DTT, pH 7.4) to the column in a linear gradient (0.1 ml/min). The proteins were then eluted using 5 ml elution buffer (20 mM HEPES, 500 mM NaCl, 500 mM imidazole, 0.5% Tween 20, and 5% glycerol, pH 7.4) and concentrated via ultrafiltration (Amicon, Millipore).
For EMSA, the purified His6-OsnR proteins (maximum of 1.2 μg) were incubated with 30 ng of DNA fragments, which were prepared using PCR withC. glutamicumgenomic DNA as the template. The primers used for the PCR amplification are listed in Additional file1: Table S2, and the resulting amplified DNA were 200–300 bp. The DNA–protein binding reaction was performed at 30 °C for 30 min in a total volume of 20 μl [20 mM HEPES, 2.5 mM MgCl2, 75 mM KCl, 0.5 mM EDTA, 2 mM DTT, 10% glycerol, 1 μg poly d(I-C), and 100 ng/μl BSA, pH 7.4]. When needed, diamide at the final concentration of 5, 10, or 20 mM, or DTT at 10, 20, or 40 mM were spiked into the protein sample. Bands were resolved via 5% polyacrylamide-gel electrophoresis, and DNA was visualized using GelRed nucleic acid stain (Biotium).
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its Additional files.
Abbreviations
- C. glutamicum:
-
Corynebacterium glutamicum
- ChIP-seq:
-
Chromatin immunoprecipitation-sequencing
- ROS:
-
Reactive oxygen species
- MSH:
-
Mycothiol
- EMSA:
-
Electrophoretic mobility shift assay
- E. coli:
-
Escherichia coli
- DTT:
-
Dithiothreitol
- IPTG:
-
Isopropyl β-d-1-thiogalactopyranoside
- ONPG:
-
O-Nitrophenyl-β-d-galactopyranoside
- ECF:
-
Extracytoplasmic function
- LB:
-
Luria–Bertani
- RT-qPCR:
-
Reverse transcription-quantitative polymerase chain reaction
References
Stackebrandt E, Rainey FA, Ward-Rainey NL. Proposal for a new hierarchic classification system,Actinobacteriaclassis nov. Int J Syst Bacteriol. 1997;47:479–91.
Leuchtenberger W, Huthmacher K, Drauz K. Biotechnological production of amino acids and derivatives: current status and prospects. Appl Microbiol Biotechnol. 2005;69:1–8.
Gibson BR, Lawrence SJ, Leclaire JPR, Powell CD, Smart KA. Yeast responses to stresses associated with industrial brewery handling. FEMS Microbiol Rev. 2007;31:535–69.
Machielsen R, van Alen-Boerrigter IJ, Koole LA, Bongers RS, Kleerebezem M, Van Hylckama Vlieg JE. Indigenous and environmental modulation of frequencies of mutation inLactobacillus plantarum.Appl Environ Microbiol. 2010;76:1587–95.
Schröder J, Tauch A. Transcriptional regulation of gene expression inCorynebacterium glutamicum: the role of global, master and local regulators in the modular and hierarchical gene regulatory network. FEMS Microbiol Rev. 2010;34:658–737.
Imlay农协。的分子机制和生理al consequences of oxidative stress: lessons from a model bacterium. Nat Rev Microbiol. 2013;11:443–54.
Ezraty B, Gennaris A, Barras F, Collet JF. Oxidative stress, protein damage and repair in bacteria. Nat Rev Microbiol. 2017;15:385–96.
Kim JS, Holmes RK. Characterization of OxyR as a negative transcriptional regulator that represses catalase production inCorynebacterium diphtheriae.PLoS ONE. 2012;7:e31709.
乾佑哉Teramoto H, M,汤川H . OxyR充当反式criptional repressor of hydrogen peroxide-inducible antioxidant genes inCorynebacterium glutamicumR. FEBS J. 2013;280:3298–312.
Hong EJ, Jeong H, Lee DS, Kim Y, Lee HS. TheahpDgene ofCorynebacterium glutamicumplays an important role in hydrogen peroxide-induced oxidative stress response. J Biochem. 2019;165:197–204.
Ung KSE, Av-Gay Y. Mycothiol-dependent mycobacterial response to oxidative stress. FEBS Lett. 2006;580:2712–6.
牛顿GL, Buchmeier N, Fahey RC。生物合成和functions of mycothiol, the unique protective thiol ofActinobacteria.Microbiol Mol Biol Rev. 2008;72:471–94.
Liu YB, Long MX, Yin YJ, Si MR, Zhang L, Lu ZQ, Wang Y, Shen XH. Physiological roles of mycothiol in detoxification and tolerance to multiple poisonous chemicals inCorynebacterium glutamicum.Arch Microbiol. 2013;195:419–29.
范的激光K,汉密尔顿CJ, Messens j .低分了-weight thiols in thiol-disulfide exchange. Antioxid Redox Signal. 2013;18:1642–53.
Patel MP, Blanchard JS. Expression, purification, and characterization ofMycobacterium tuberculosismycothione reductase. Biochemistry. 1999;38:11827–33.
Si M, Long M, Chaudhry MT, Xu Y, Zhang P, Zhang L, Shen X. Functional characterization ofCorynebacterium glutamicummycothiol S-conjugate amidase. PLoS ONE. 2014;9:e115075.
Si M, Zhao C, Zhang B, Wei D, Chen K, Yang X, Xiao H, Shen X. Overexpression of mycothiol disulfide reductase enhancesCorynebacterium glutamicumrobustness by modulating cellular redox homeostasis and antioxidant proteins under oxidative stress. Sci Rep. 2016;6:1–14.
Holmgren A. Thioredoxin. Ann Rev Biochem. 1985;54:237–71.
Prinz WA, Åslund F, Holmgren A, Beckwith J. The role of the thioredoxin and glutaredoxin pathways in reducing protein disulfide bonds in theEscherichia colicytoplasm. J Biol Chem. 1997;272:15661–7.
Powis G, Montfort WR. Properties and biological activities of thioredoxins. Annu Rev Biophys Biomol Struct. 2001;30:421–55.
Mailloux RJ, Harper ME. Uncoupling proteins and the control of mitochondrial reactive oxygen species production. Free Radic Biol Med. 2011;51:1106–15.
Kim TH, Park JS, Kim HJ, Kim Y, Kim P, Lee HS. ThewhcEgene ofCorynebacterium glutamicumis important for survival following heat and oxidative stress. Biochem Biophys Res Commun. 2005;337:757–64.
Choi WW, Park SD, Lee SM, Kim HB, Kim Y, Lee HS. ThewhcAgene plays a negative role in oxidative stress response ofCorynebacterium glutamicum.FEMS Microbiol Lett. 2009;290:32–8.
Pedre B, Young D, Charlier D, Mourenza Á, Rosado LA, Marcos-Pascual L, Wahni K, Martens E, de la Rubia AG, Belousov VV, Mateos LM, Messens J. Structural snapshots of OxyR reveal the peroxidatic mechanism of H2O2sensing. Proc Natl Acad Sci USA. 2018;115:E11623–32.
Si M, Su T, Chen C, Liu J, Gong Z, Che C, Li G, Yang G. OhsR acts as an organic peroxide-sensing transcriptional activator using asS-mycothiolation mechanism inCorynebacterium glutamicum.Microb Cell Fact. 2018;17:200.
Bussmann M, Baumgart M, Bott M. RosR (Cg1324), a hydrogen peroxide-sensitive MarR-type transcriptional regulator ofCorynebacterium glutamicum.J Biol Chem. 2010;285:29305–18.
Si M, Chen C, Zhong J, Li X, Liu Y, Su T, Yang G. MsrR is a thiol-based oxidation-sensing regulator of the XRE family that modulatesC. glutamicumoxidative stress resistance. Microb Cell Fact. 2020;19:189.
Si M, Chen C, Su T, Che C, Yao S, Liang G, Li G, Yang G. CosR is an oxidative stress sensing a MarR-type transcriptional repressor inCorynebacterium glutamicum.Biochem J. 2018;475:3979–95.
Ehira年代,荻H, Teramoto H, Iuni M,汤川H .再保险gulation of quinone oxidoreductase by the redox-sensing transcriptional regulator QorR inCorynebacterium glutamicum.J Biol Chem. 2009;284:16736–42.
Si M, Chen C, Che C, Liu Y, Li X, Su T. The thiol oxidation-based sensing and regulation mechanism for the OasR-mediated organic peroxide and antibiotic resistance inC. glutamicum.Biochem J. 2020;477:3709–27.
Hong EJ, Kim P, Kim ES, Kim Y, Lee HS. Involvement of theosrRgene in the hydrogen peroxide-mediated stress response ofCorynebacterium glutamicum.Res Microbiol. 2016;167:20–8.
Kim TH, Kim HJ, ParK JS, Kim Y, Kim P, Lee HS. Functional analysis ofsigHexpression inCorynebacterium glutamicum.Biochem Biophys Res Commun. 2005;331:1542–7.
Busche T, Šilar R, Pičmanová M, Pátek M, Kalinowski J. Transcriptional regulation of the operon encoding stress-responsive ECF sigma factor SigH and its anti-sigma factor RshA, and control of its regulatory network inCorynebacterium glutamiucm.BMC Genom. 2012;13:445.
Kohl TA, Tauch A. The GlxR regulon of the amino acid producerCorynebacterium glutamicum: detection of the corynebacterial core regulon and integration into the transcriptional regulatory network model. J Biotechnol. 2009;143:239–46.
Schröder J, Tauch A. Transcriptional regulation of gene expression inCorynebacterium glutamicum: the role of global, master and local regulators in the modular and hierarchical gene regulatory network. FEMS Microbiol Rev. 2010;34:685–737.
Jeong H, Kim Y, Lee HS. TheosnRgene ofCorynebacterium glutamicum在氧化压力起着负监管的作用ss responses. J Ind Microbiol Biotechnol. 2019;46:241–8.
Baulard AR, Betts JC, Engohang-Ndong J, Quan S, McAdam RA, Brennan PJ, Locht C, Besra GS. Activation of the pro-drug ethionamide is regulated in mycobacteria. J Biol Chem. 2000;275:28326–31.
Vilchèze C, Baughn AD, Tufariello J, Leung LW, Kuo M, Basler CF, Alland D, Sacchettini JC, Freundlich JS, Jacobs WR Jr. Novel inhibitors of InhA efficiently killMycobacterium tuberculosisunder aerobic and anaerobic conditions. Antimicrob Agents Chemother. 2011;55:3889–98.
Reardon-Robinson ME, Ton-That H. Disulfide-bond-forming pathways in Gram-positive bacteria. J Bacteriol. 2016;198:746–54.
Pfeifer-Sancar K, Mentz A, Rückert C, Kalinowski J. Comprehensive analysis of theCorynebacterium glutamicumtranscriptome using an improved RNAseq technique. BMC Genom. 2013;14:888.
Toyoda K, Inui M. Regulons of global transcription factors inCorynebacterium glutamicum.Appl Microbiol Biotechnol. 2016;100:45–60.
Dostálová H, Holátko J, Busche T, Rucká L, Rapoport A, Halada P, Nešvera J, Kalinowski J, Pátek M. Assignment of sigma factors of RNA polymerase to promoters inCorynebacterium glutamicum.AMB Express. 2017;7:133.
Chowdhury RP, Gupta S, Chatterji D. Identification and characterization of thedpspromoter ofMycobacterium smegmatis: promoter recognition by stress-specific extracytoplasmic function sigma factors σHand σF.J Bacterial. 2007;189:8973–81.
Yanamandra SS, Sarrafee SS, Anaya-Bergmann C, Jones K, Lewis JP. Role of thePorphyromonas gingivalisECF sigma factor. SigH Mol Oral Microbiol. 2012;27:202–19.
Pátek M, Nešvera J. Sigma factors and promoters inCorynebacterium glutamicum.J Biotechnol. 2011;154:101–13.
Van Bergen LAH, Roos G, De Proft F. From thiol to sulfonic acid: modeling the oxidation pathway of protein thiols by hydrogen peroxide. J Phys Chem A. 2014;118:6078–84.
Rosenfeld N, Elowitz MB, Alon U. Negative autoregulation speeds the response times of transcription networks. J Mol Biol. 2002;323:785–93.
Kumar S, Badireddy S, Pal K, Sharma S, Arora C, Garg SK, Alam MS, Agrawal P, Anand GS, Swaminathan K. Interaction ofMycobacterium tuberculosisRshA and SigH is mediated by salt bridges. PLoS ONE. 2012;7:e43676.
Lee C, Lee SM, Mukhopadhyay P, Kim SJ, Lee SC, Ahn WS, Yu MH, Storz G, Ryu SE. Redox regulation of OxyR requires specific disulfide bond formation involving a rapid kinetic reaction path. Nat Struct Mol Biol. 2004;11:1179–85.
Klomsiri C, Karplus PA, Poole LB. Cysteine-based redox switches in enzymes. Antioxid Redox Signal. 2011;14:1065–77.
Hillion M, Antelmann H. Thiol-based redox switches in prokaryotes. Biol Chem. 2015;396:415–44.
Rodrigue S, Brodeur J, Jacques PÉ, Gervais AL, Brzezinski R, Gaudreau L. Identification of Mycobacterial σ factor binding sites by chromatin immunoprecipitation assays. J Bacteriol. 2007;189:1505–13.
Fitzgerald DM, Bonocora RP, Wade JT. Comprehensive mapping of theEscherichia coliflagellar regulatory network. PLoS Genet. 2014;10:e1004649.
Minch KJ, Rustad TR, Peterson EJR, Winkler J, Reiss DJ, Ma S, Hickey M, Brabant W, Morrison B, Turkarslan S, Mawhinney C, Galagan JE, Price ND, Baliga NS, Sherman DR. The DNA-binding network ofMycobacterium tuberculosis.Nat Commun. 2015;6:5829.
Sharp JD, Singh AK, Park ST, Lyubetskaya A, Peterson MW, Gomes ALC, Potluri LP, Raman S, Galagan JE, Husson RN. Comprehensive definition of the SigH regulon ofMycobacterium tuberculosisreveals transcriptional control of diverse stress responses. PLoS ONE. 2016;11(3):e0152145.
Toyoda K, Teramoto H, Yukawa H, Inui M. Expanding the regulatory network governed by the extracytoplasmic function sigma factor σHinCorynebacterium glutamicum.J Bacteriol. 2015;197:483–96.
Ehira S, Teramoto H, Iuni M, Yukawa H. Regulation ofCorynebacterium glutamicumheat shock response by the extracytoplasmic-function sigma factor SigH and transcriptional regulators HspR and HrcA. J Bacteriol. 2009;191:2964–72.
Chi BK, Busche T, Laer KV, Bäsell K, Becher D, Clermont L, Seibold GM, Persicke M, Kalinowski J, Messens J, Antelmann H. ProteinS-mycothiolation functions as redox-switch and thiol protection mechanism inCorynebacterium glutamicumunder hypochlorite stress. Antioxid Redox Signal. 2014;20:589–605.
Vetting MW, Frantom PA, Blanchard JS. Structural and enzymatic analysis of MshA fromCorynebacterium glutamicum: substrate-assisted catalysis. J Biol Chem. 2008;283:15834–44.
Liu YB, Chen C, Chaudhary MT, Si MR, Zhang L, Wang Y, Shen XH. EnhancingCorynebacterium glutamicumrobustness by over-expressing a gene,mshA, for mycothiol glycosyltransferase. Biotechnol Lett. 2014;36:1453–9.
Dostálová H, Busche T, Holátko J, Rucká L, Štěpánek V, Barvík I, Nešvera J, Kalinowski J, Pátek M. Overlap of promoter recognition specificity of stress response sigma factors SigD and SigH inCorynebacterium glutamicumATCC 13032. Front Microbiol. 2019;9:3287.
Martin JL, Bardwell JCA, Kuriyan J. Crystal structure of the DsbA protein required for disulphide bond formationin vivo.Nature. 1993;365:464–8.
Missiakas D, Georgopoulos C, Raina S. Identification and characterization of theEscherichia coligenedsbB, whose product is involved in the formation of disulfide bondsin vivo.Proc Natl Acad Sci USA. 1993;90:7084–8.
Bardwell JC, Lee JO, Jander G, Martin N, Belin D, Beckwith J. A pathway for disulfide bond formationin vivo.Proc Natl Acad Sci USA. 1993;90:1038–42.
Dutton RJ, Boyd D, Berkmen M, Beckwith J. Bacterial species exhibit diversity in their mechanisms and capacity for protein disulfide bond formation. Proc Natl Acad Sci USA. 2008;105:11933–8.
Daniels R, Mellroth P, Bernsel A, Neiers F, Normark S, von Heijne G, Henriques-Normark B. Disulfide bond formation and cysteine exclusion in gram-positive bacteria. J Biol Chem. 2010;285:3300–9.
Follettie MT, Peoples OP, Agoropoulou C, Sinskey AJ. Gene structure and expression of theCorynebacterium glutamicumN13ask-asdoperon. J Bacteriol. 1993;175:4096–103.
Sambrook J, Fritsch EF, Maniatis R. Molecular cloning: a laboratory manual. 3rd ed. New York: Cold Spring Harbor Laboratory; 2001.
Park SD, Lee SN, Park IH, Choi JS, Jeong WK, Kim Y, Lee HS. Isolation and characterization of transcriptional elements fromCorynebacterium glutamicum.生物科技Microbiol》。2004;14:789 - 95。
Simon RV, Houman F, Kleckner N. Improved single and multicopylac-based cloning vectors for protein and operon fusions. Gene. 1987;53:85–96.
Jeong H, Lee JH, Kim Y, Lee HS. Thiol-specific oxidant diamide downregulateswhiAgene ofCorynebacterium glutamicum, thereby suppressing cell division and metabolism. Res Microbiol. 2020;171:331–40.
Miller JH. Experiments in molecular genetics. New York: Cold Spring Harbor; 1972.
Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54.
Jungwirth B, Sala C, Kohl TA, Uplekar S, Baumbach J, Cole ST, Pühler A, Tauch A. High-resolution detection of DNA binding sites of the global transcriptional regulator GlxR inCorynebacterium glutamicum.Microbiology. 2013;159:12–22.
Acknowledgements
The authors wish to thank Dr. Jung Chul Park (currently at Samsung Advanced Institute of Technology) for technical assistance in preparing the vectors used in the in vivo analyses.
Funding
This research was financially supported by the Basic Science Research Program, through the National Research Foundation of Korea (NRF), funded by the Ministry of Education (2019R1I1A3A01051388), and in part by a grant from Korea University.
Author information
Authors and Affiliations
Contributions
HJ conducted the experiments, analyzed the data, and drafted the manuscript. YK conceptualized the study, and critically read and finalized the manuscript. HSL supervised the study and procured the funding. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
Bacterial strains and plasmids used in this study.Table S2.Oligonucleotides used in this study.Figure S1.Binding of the purified OsnR protein on the promoter regions of thetrxBandsodAgenes.Figure S2.The mRNA levels of genes linked to iron homeostasis inC. glutamicumcells.Figure S3.Transcription of themyc-osnRfusion gene as measured by qRT-PCR.C. glutamicumcells were grown in minimal media.
Rights and permissions
Open AccessThis article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visithttp://creativecommons.org/licenses/by/4.0/.The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Jeong, H., Kim, Y. & Lee, HS. OsnR is an autoregulatory negative transcription factor controlling redox-dependent stress responses inCorynebacterium glutamicum.Microb Cell Fact20, 203 (2021). https://doi.org/10.1186/s12934-021-01693-1
Received:
Accepted:
Published:
DOI:https://doi.org/10.1186/s12934-021-01693-1